
- DOWNLOAD CYTOSCAPE REGISTRATION
- DOWNLOAD CYTOSCAPE SOFTWARE
- DOWNLOAD CYTOSCAPE CODE
- DOWNLOAD CYTOSCAPE DOWNLOAD
Thus, plug-ins also provide a general means of introducing and testing new features. Several of Cytoscape's core features (graph layout and node attribute mapping) ultimately led us to incorporate it into theĬytoscape Core platform. ↵ 7 Although attribute-based layout was initially implemented as a plug-in, its general applicability and tight integration with Click 'SSH and GPG keys', then follow the instructions on the linked guide to generate an SSH key on your particular operating system. Then, click the arrow in the top-right hand corner of any GitHub page and choose 'Settings'.
To accommodate new biological problems-is gained by not inscribing the notion of specific biological entities directly into To do this, you will first need to be added to the Cytoscape GitHub project by one of the core developers. Node attributes entitled “expression ratio” or “cellular compartment.” In this way, great freedom and flexibility-the ability For example, the core may represent a node (an abstract conceptįree of biological semantics) whose label is GCN4 (a text string with significance to yeast biologists as an important transcription factor) or we might use the Core to define Of course, there is often substantialīiological significance associated with data in the Core. Problem by leaving it to plug-in writers to adopt semantics adequate to the problem at hand. Were in the Cytoscape Core, we would be faced with a difficult question which semantics should we use? Cytoscape avoids this ↵ 6 Biological semantics vary widely within the biological community as well as fromproject to project.
DOWNLOAD CYTOSCAPE DOWNLOAD
[The Cytoscape v1.1 Core runs on all major operating systems and is freely available for download from Interaction network for Halobacterium, and an interface to detailed stochastic/kinetic gene regulatory models. SeveralĬase studies of Cytoscape plug-ins are surveyed, including a search for interaction pathways correlating with changes in geneĮxpression, a study of protein complexes involved in cellular recovery to DNA damage, inference of a combined physical/functional The Core is extensible throughĪ straightforward plug-in architecture, allowing rapid development of additional computational analyses and features.
DOWNLOAD CYTOSCAPE SOFTWARE
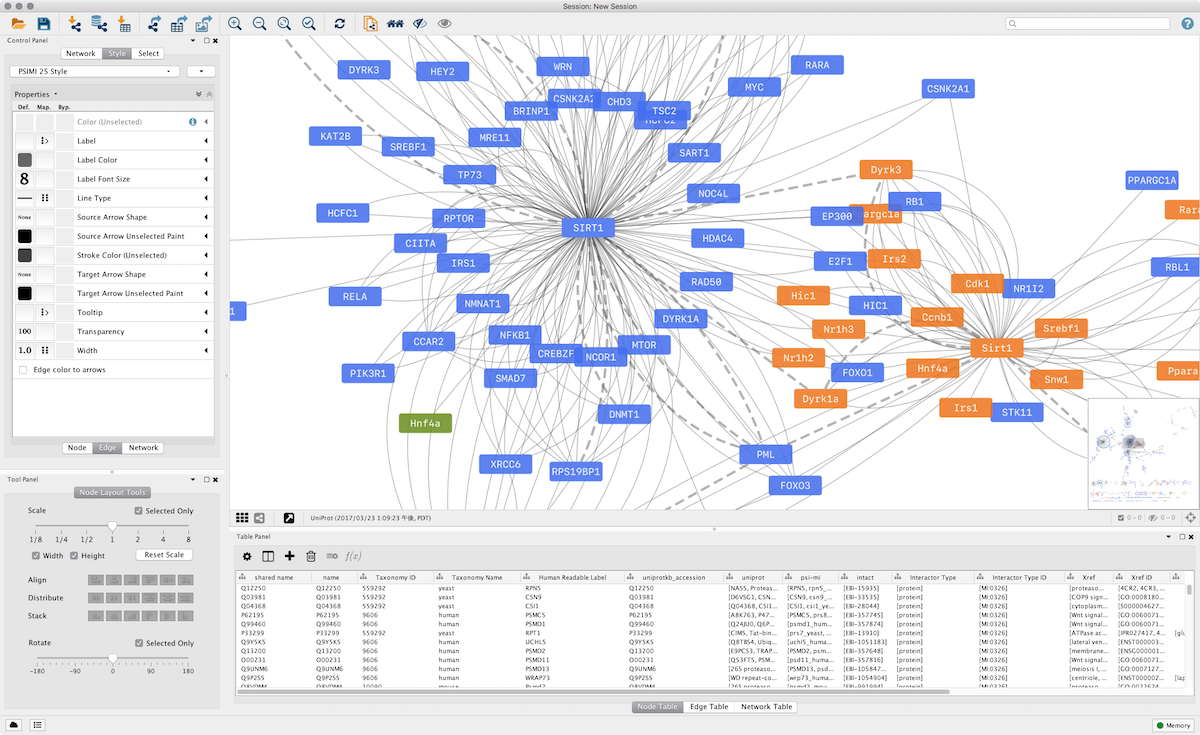
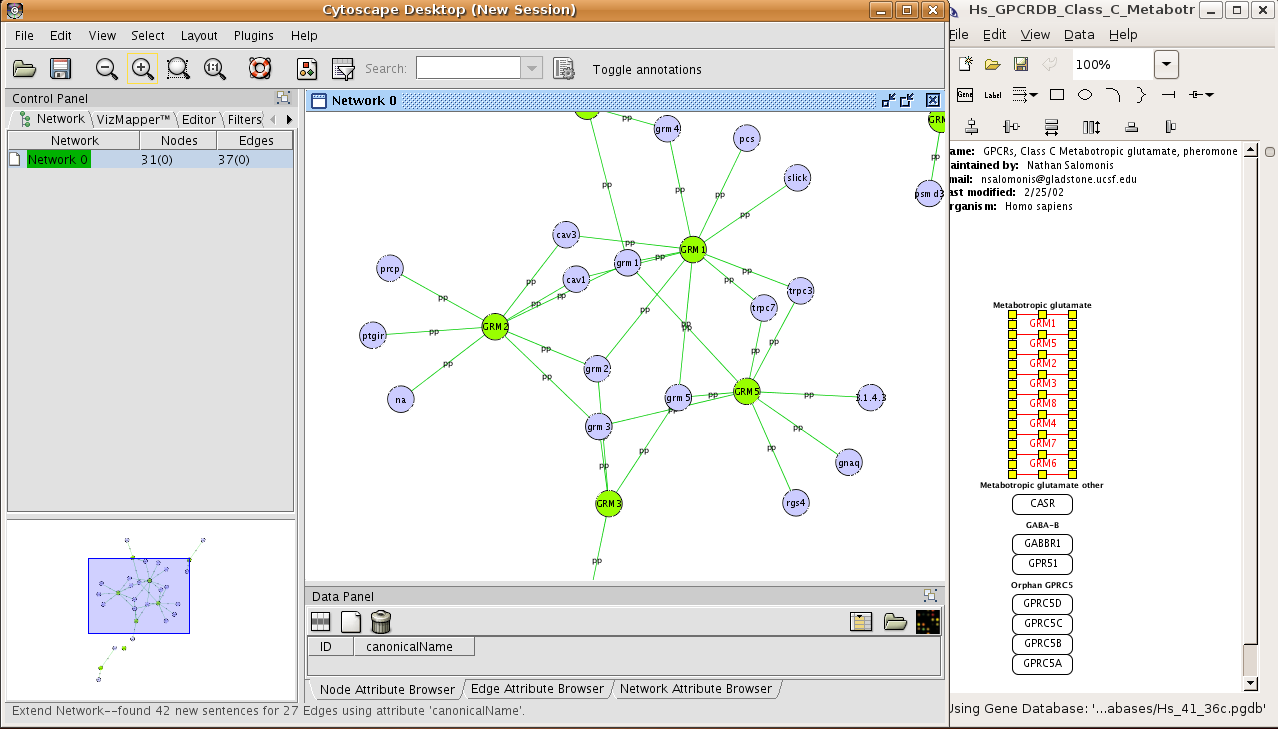
Cytoscape's software Core providesīasic functionality to layout and query the network to visually integrate the network with expression profiles, phenotypes,Īnd other molecular states and to link the network to databases of functional annotations. Although applicable to any system of molecular componentsĪnd interactions, Cytoscape is most powerful when used in conjunction with large databases of protein-protein, protein-DNA,Īnd genetic interactions that are increasingly available for humans and model organisms. Lastly, the tool has strong support for system biology, proteomics, and genomics.Cytoscape is an open source software project for integrating biomolecular interaction networks with high-throughput expressionĭata and other molecular states into a unified conceptual framework.
DOWNLOAD CYTOSCAPE REGISTRATION
Its important to notice that Cytoscape requires registration to download the program. It should be noted that while its interface is nice and well-structured, it will take some time for an average user to understand its functions and layout. Cytoscape is an open source software platform for visualizing data. Download 3.9. This software platform is a very powerful one created to deal with both simple and complex tasks. These plugins can be used for network and a lot more things like molecular profiling analyses, additional file format support, new layouts, searching in large networks, connection with databases just to mention a few. It comes with robust functionality that boasts of many additional features that are available in the form of plugins. It is an open source bioinformatics software utility that visualizes molecular interaction networks and also integrates with gene expression profiles as well as other state data. Cytoscape is available as a platform-independent open-source Java application, released under the terms of the LGPL. Whether you’re a student, teacher, lecturer or just a mere enthusiast in this field, this program is one you should definitely try out.
DOWNLOAD CYTOSCAPE CODE
If you are the type with a knack for biology and other related aspects of life, you definitely need a tool to further enhance your studies, research, and findings. Cytoscape Web » Download Contents Show all sections Version 1.0.4 (latest) Version 1.0.3 Version 1.0.2 Source Code Version 1.0.4 (latest) Download This minor release fixes a bug that prevented loops (edges that connect a node to itself) from being rendered.


 0 kommentar(er)
0 kommentar(er)
